Broader Goal of our lab
We all start life as a single cell. The process of developing from a single cell to an organism requires the precise coordination of tens of thousands of coding genes in space and time to gives rise to a myriad of cell types. This coordinated gene regulation is dictated by distal-regulatory elements called “enhancers.” Enhancers locate their cognate promoter in the dense 3D space of the nucleus and deliver transcriptional machinery to promoters for their activation. They act as a single unit (singleton enhancer) or in a group (clustered enhancers) to activate target genes. Additionally, active enhancers transcribe enhancer-RNAs (eRNAs) whose functions are not known. The prevailing model suggests that an enhancer’s search for a promoter is restricted within a local chromatin domain known as a Topologically Associating Domain (TAD). However, the functional link between enhancer mediated gene expression and chromatin topology is poorly understood.
The goal of our research is to extend the understanding of these molecular mechanisms underlying the linkages between chromatin domains, distal regulatory elements and associated non-coding RNAs (the enhancer RNAs) in gene regulation. The gained knowledge will help develop strategies to target enhancer to amend the mis-expression of disease related causal genes as “enhancer therapy.” We use combination of approaches such as Genetic perturbations, molecular biology, biochemistry, genome sequencing and live cell imaging to functionally tie the enhancers, transcription and genome folding.
Specific Projects
The ligand-induced enhancer clusters are pre-seeded under basal signaling:
How transcriptional response to cyclic hormonal signaling is so reproducible (estrogen cycles during menstruation and glucocorticoids peaks every 24h). We hypothesised that since ligand-dependent transcription is driven by enhancers perhaps, these enhancers could carry a memory (bookmarking) bet
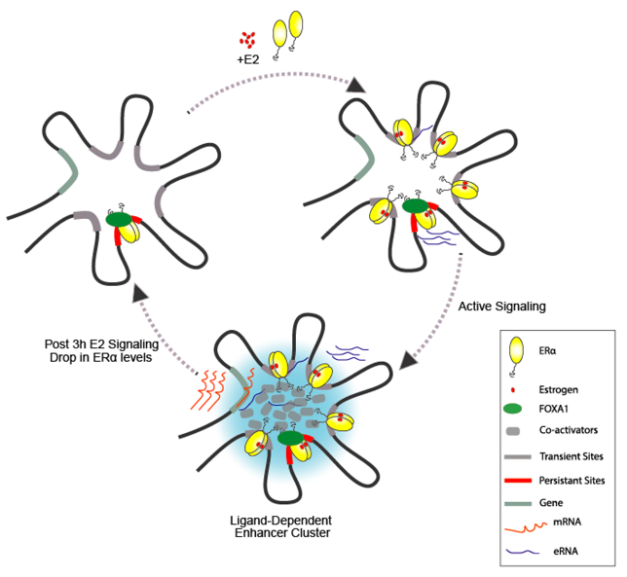
ween each cycle of ligand stimulation ensuring faithful and noise-free signaling response.
Interrogating enhancer memory, our group recently showed that upon estrogen stimulation, the ERα binding in the genome is clustered where one lead enhancer in the cluster is marked (pre-seeded) by an unliganded ERα-receptor even before signaling. These clusters emerge upon signaling and though they disappear at the end of signaling, they leave behind the same pre-marked enhancer still bound by the receptor for next round of ligand exposure. These enhancer clusters are identified as phase separated ERα-condensates and any perturbation in pre-seeding abolishes the enhancer-cluster formation and therefore, the transcription of genes and also their periodicity. We are now testing the extent of enhancer bookmarking in other signalling systems including hormones such as androgens and glucocorticoids. (Saravanan et al., 2020), (Blobel et al., 2021), (Mann & Notani, 2023).
Enhancer clusters do not act in sum-of-all manner:
Interestingly, though ERα enhancer clusters regulate expression of estrogen target genes but they do not exhibit the features of canonical super-enhancers (high levels of Mediator, prescribed distance between enhancers). Furthermore, as opposed to conventional thinking that super enhancers regulate target gene as a sum of all enhancers, ERα clustered enhancers exhibited sole dependence on bookmarked enhancers. These observations prompted us to functionally dissect the relationship of individual enhancers in super enhancers.
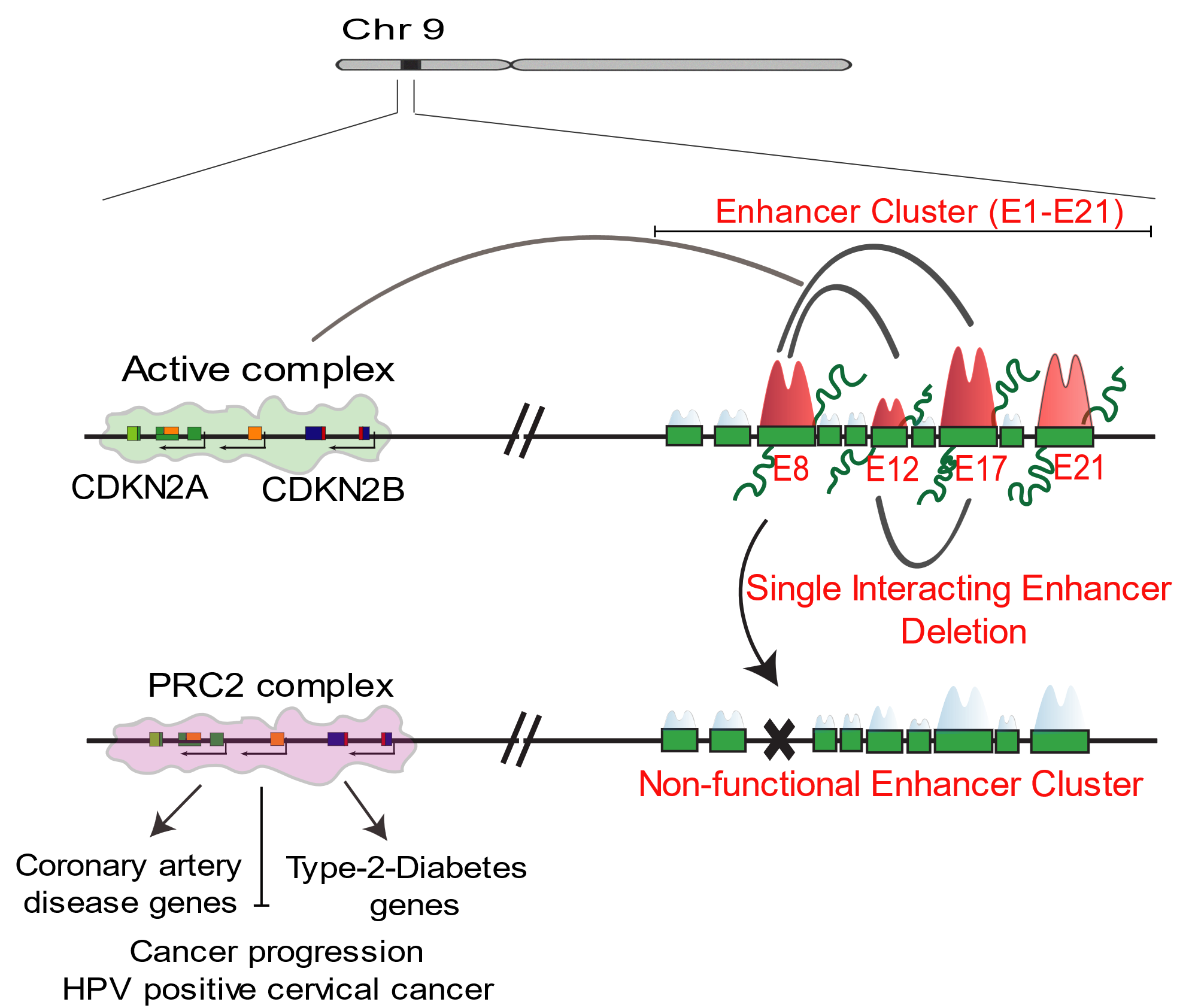
We tested these concepts on one of the most dense-enhancer clusters in the genome at the 9p21 locus which is a GWAS hotspot. We observe that not all enhancers within a cluster are functional and that functional enhancers form an interdependent network where they rely on each other for a net effect on target gene transcription. Furthermore, not all functional enhancers are biochemically marked with high levels of enhancer marks.
By functionally dissecting enhancer cluster at 9p21 locus and, signaling driven transient enhancer cluster, we reveal the unexpected dependence of enhancer clusters on single to few functional enhancers emphasising that the effect of super enhancer on target genes is not the sum of constituent enhancers. Our studies underscore the inadequacy of super enhancer based assigning of functionality as currently practiced and emphasises that, even though laborious, a careful enhancer-by-enhancer dissection of SE function is the only way forward towards a comprehensive understanding of SE functional diversity. (Farooq, et al., 2021).
Do enhancers contribute to genome organization:
While dissecting the hierarchy of enhancers within the clusters at the 9p21 locus,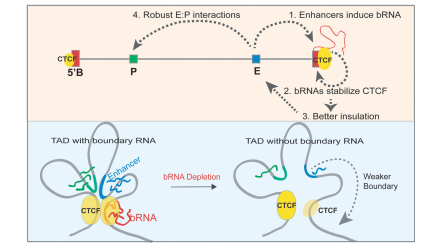 we discovered that functional enhancers directly assist TAD boundaries to recruit CTCF (a transcription factor present at most TAD boundaries and its interactions with cohesin complex organizes TADs). Briefly, as the enhancers become active during signaling/development, they loop with boundaries of the same TAD. This looping results in the transcription of non-coding RNAs at the boundaries. These RNAs then interact with CTCF to stabilize its binding at the TAD borders. As a result, these TADs are better insulated and do not allow the enhancers to interact with genes outside the given TAD. (Islam, et al., 2023)
we discovered that functional enhancers directly assist TAD boundaries to recruit CTCF (a transcription factor present at most TAD boundaries and its interactions with cohesin complex organizes TADs). Briefly, as the enhancers become active during signaling/development, they loop with boundaries of the same TAD. This looping results in the transcription of non-coding RNAs at the boundaries. These RNAs then interact with CTCF to stabilize its binding at the TAD borders. As a result, these TADs are better insulated and do not allow the enhancers to interact with genes outside the given TAD. (Islam, et al., 2023)
Understanding enhancer functions through disease-causing mutations:
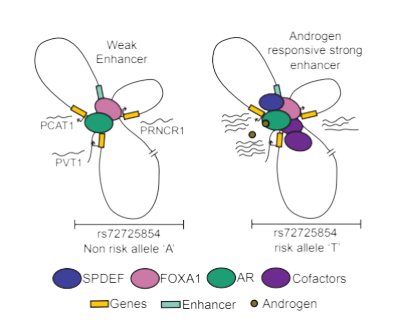
In addition, I have previously investigated how mutations within the enhancer at 9p21 gene desert enhance the susceptibility of a population to coronary artery disease (CAD). We used the same concept recently to understand why men of African ancestry are more predisposed to prostate cancer. For the first time, we have shown that even the rare genetic variants can be causal as they can create a hormone-sensitive stronger enhancer that in turn can upregulate pro-oncogenic lncRNAs to enhance prostate tumor susceptibility in young men of African ancestry. (Kaiwalaya, et al., 2020).

