Prof. Upinder S. Bhalla - Research
| 1.Sequence computations in the brain (Bhanu Priya Somashekar, Upi Bhalla) |
| 2. Sequence and EI computations in the brain(Aditya Asopa, Anzal KS, Sulu Mohan) |
| 3. Sequence computations in the brain: in-vivo work (Soumya Bhattacharjee, Hrishikesh Nambisan ) |
| 4. Signaling and physiology in health and disease: models and slice (NA Viswan, G.V. HarshaRani, Anal Kumar, Vinu Pulikkottil) |
| 5. Tool development (G.V. Harsha Rani, Upi Bhalla) |
1. Sequence computations in the brain: [ Model ] (Bhanu Priya Somashekar, Dilawar Singh, Sathyaa Subramaniam, Upi Bhalla)
We study sequence computations using theory and simulations at subcellular, neuronal, and network scales. We recently showed how single neurons can recognize when sequential input arrives on a dendrite, ordered both in time, and in space (Bhalla, eLife, 2017 doi: 10.1002/hipo.22806).
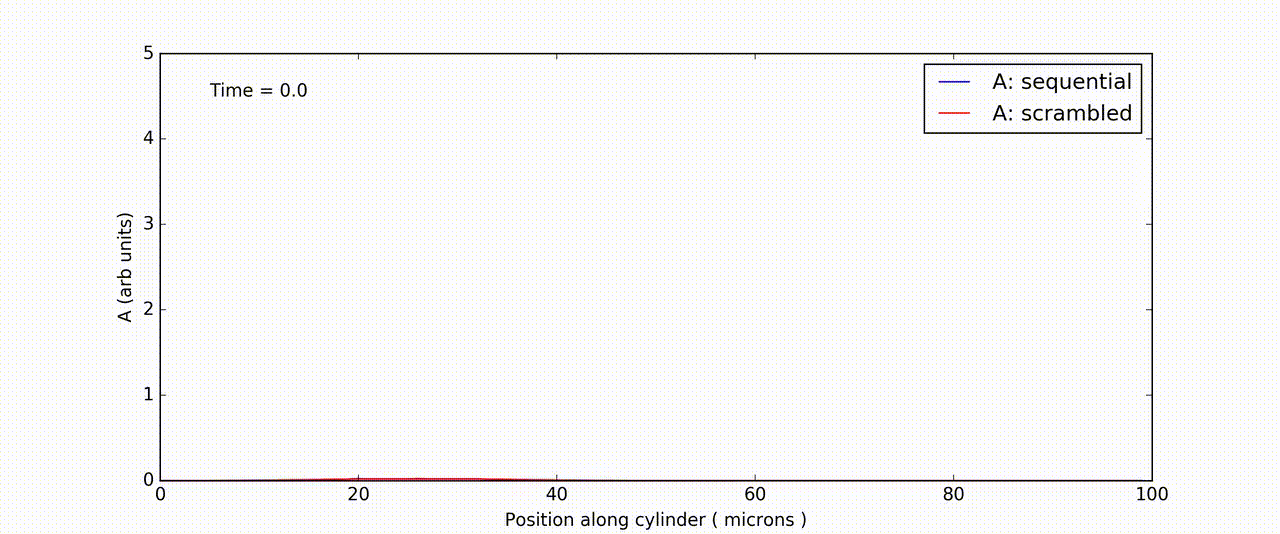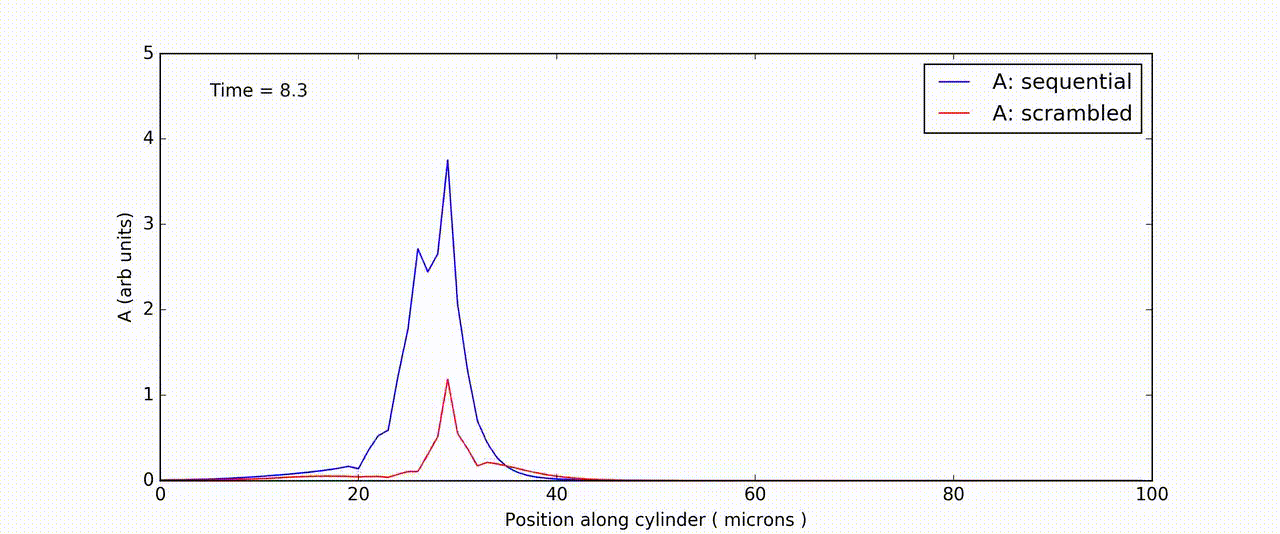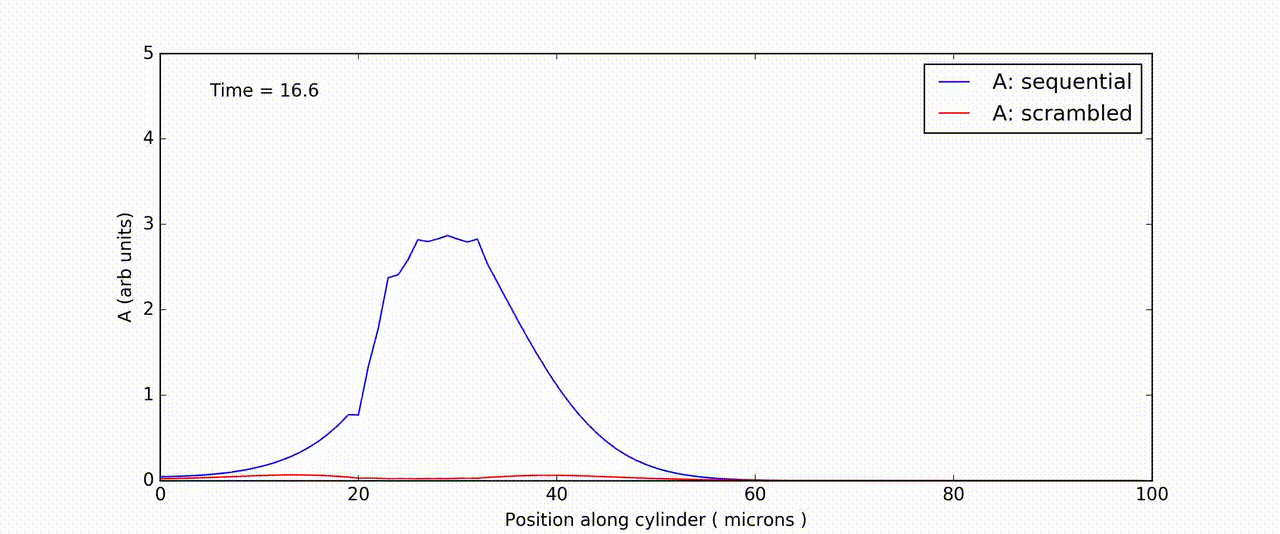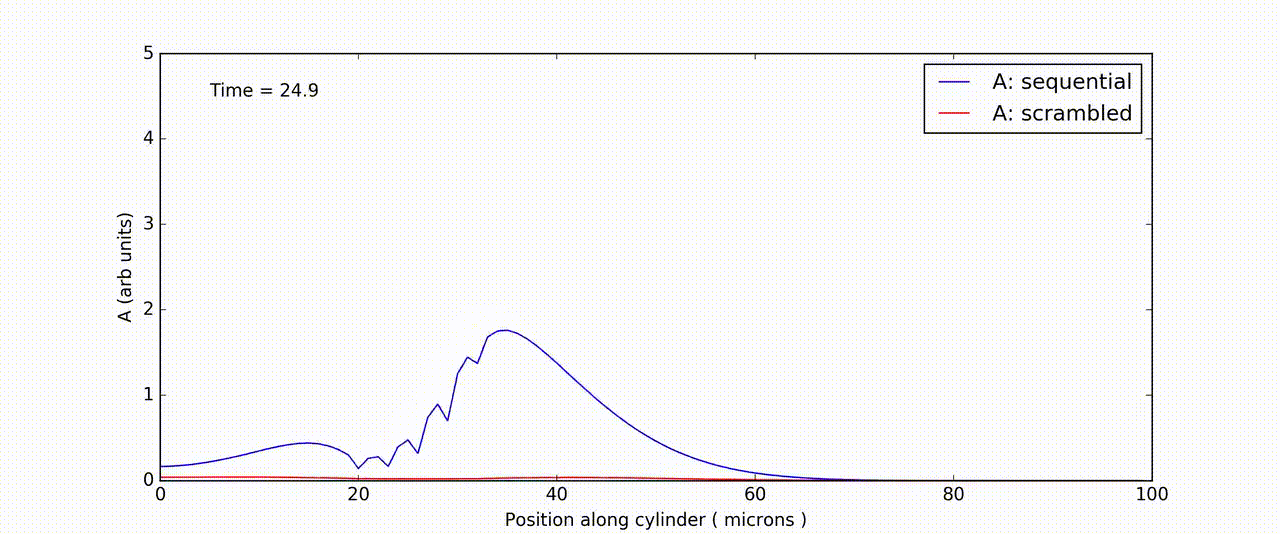
Figure: Time-and-space series of response of FitzHuge-Nagumo model to sequential (blue) and scrambled (red) input.
https://doi.org/10.7554/eLife.25827.006
We are digging deeper into the mechanisms for such sequential recognition, with special interest in calcium-induced calcium release (CICR) as a mechanism. With this there are specific implications for the timing and the spatial extent of such processes. We are then asking how such sequence-recognizing neurons might function in a network. How, for example, could the wiring for such processes be set up? Finally, we are scrutinizing the link between sequences, habituation, and novelty recognition. We find that these concepts may be closely related on all the scales we study: subcellular, neuronal and network.
2. Sequence and EI computations in the brain: [ Slice Experiments + theory ] (Aanchal Bhatia, Sahil Moza, Aditya Asopa, Anzal KS, Sulu Mohan, Collaboration with Arvind Kumar and Shreya Lakhera KTH)
We are keen to map our models of neuronal sequences to hard data. We use optogenetic methods to stimulate brain slices from the hippocampus, which is a region of the brain specially implicated in learning and memory. We deliver spatiotemporal light stimuli - basically movies - to illuminate cells in desired patterns. In one set of projects we have analyzed how inputs are summed and balanced between excitatory and inhibitory pathways. We find that there is very precise balance, both in amplitude and time. Collectively this generates an effective mechanism for network gain control as well as selective response to particular inputs out of the thousands arriving at a cell (Bhatia A, Moza S, Bhalla US, eLife 2019, doi: 10.7554/eLife.43415) Based on theoretical work from the Kumar lab (doi: https://doi.org/10.1101/428649), we are investigating the role and prevalence of asymmetry in functional responses in the hippocampus. We use similar optical stimuli to find if responses have the kinds of asymmetry that theory predicts may lead to generation of sequences. In a new set of experiments, we are developing approaches to delivering sequential input in the slice to directly test our models of single-cell ability to recognize sequences.
3. Sequence computations in the brain: [ in-vivo work ] (Soumya Bhattacharjee, Kambadur Ananthamurthy, Hrishikesh Nambisan )
We place little glass windows on the heads of mice so that we can image their brain activity while they learn. We use genetically modified mice, whose brain cells change fluorescence (flash) when the cells are active. This can be picked up using our custom-built 2-photon microscope. We train these mice on a range of tasks to associate a sound or light with a puff of air that makes them blink. As they learn to blink _before_ the air-puff, we find that activity sequences arise in their hippocampus. (Modi MN et al Elife 2014 doi: 10.7554/eLife.01982 ) We are analyzing how these sequences depend on stimuli, timing, and stages of learning.
4. Signaling and physiology in health and disease: [ Models and slice ] (Nisha NA, G. V. HarshaRani, Surbhit Wagle, Deepanjali Dwivedi, Anal Kumar, Vinu Pulikkottil, Vinod Ugale: visiting scholar)
We develop and refine very detailed models of cellular signaling and electrophysiology, with particular interest in neuronal diseases such as Autism. This is now part of a growing consortium effort to understand these conditions (SANKET/AutSim ).
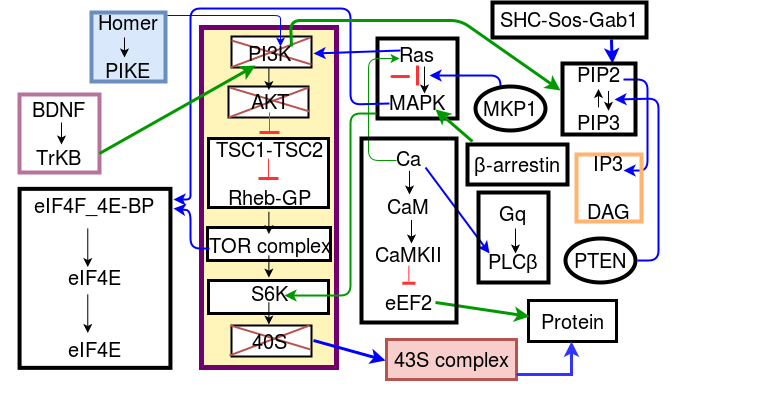
We have compiled a database of almost 200 experiments codified in standarized templates so that we can run any experiment against our models, and use various numerical methods to improve the model accuracy. We also perform slice recordings from mutant mice having a knockout in the Fragile X protein, which is implicated in autism. We use these recordings to identify what goes wrong in the cell. Further we model the properties of healthy and diseased cells so as to better understand how functional differences, such as variability, emerge from the mutations. We also model the chemical and mechanical signaling that occurs when new synaptic spines emerge on neuronal dendrites, as this is a crucial step in memory formation.
Collaborators:
Ravi Iyengar, Mount Sinai School of Medicine, New York, USA,
Dinu Florin Albeanu, Cold Spring Harbor Laboratory,
Venki Murthy, Harvard,
Naren Ramakrishnan, Virginia Tech
Raphael Camargo, Brazil
Matthias Hennig,University of Edinburgh
Jeanette Hellgren-Kotaleski and
Mikael Djurfeldt: KTH Stockholm
<a data-cke-saved-href="https://qbio.umontpellier.fr/team/ovidiu-radulescu/" href="https://qbio.umontpellier.fr/team/ovidiu-radulescu/" target="_" blank"="">



