Prof. R. Sowdhamini - Pretty Pictures
1. Domain arrangements in the Protein a-transducin
 |
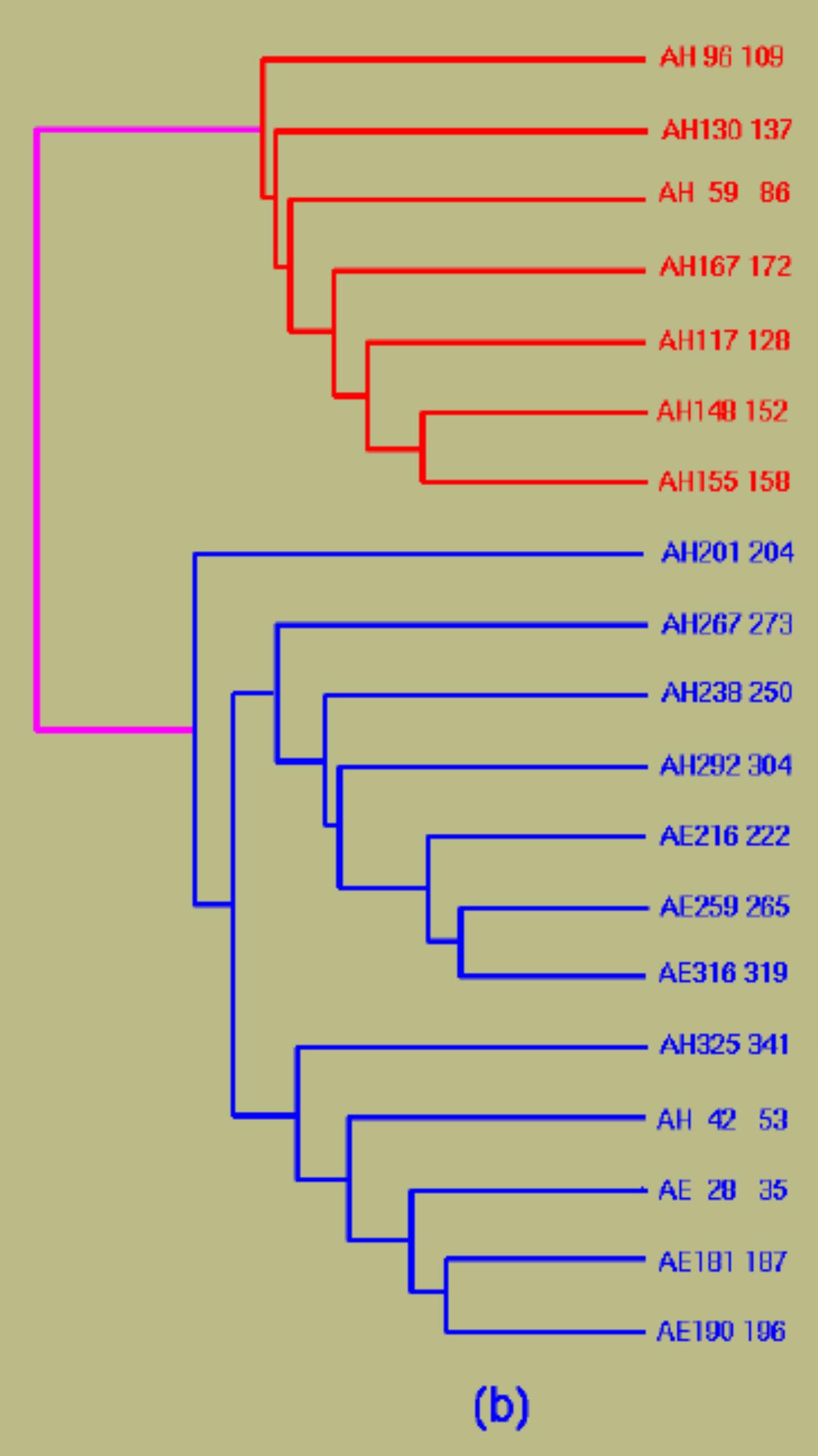 |
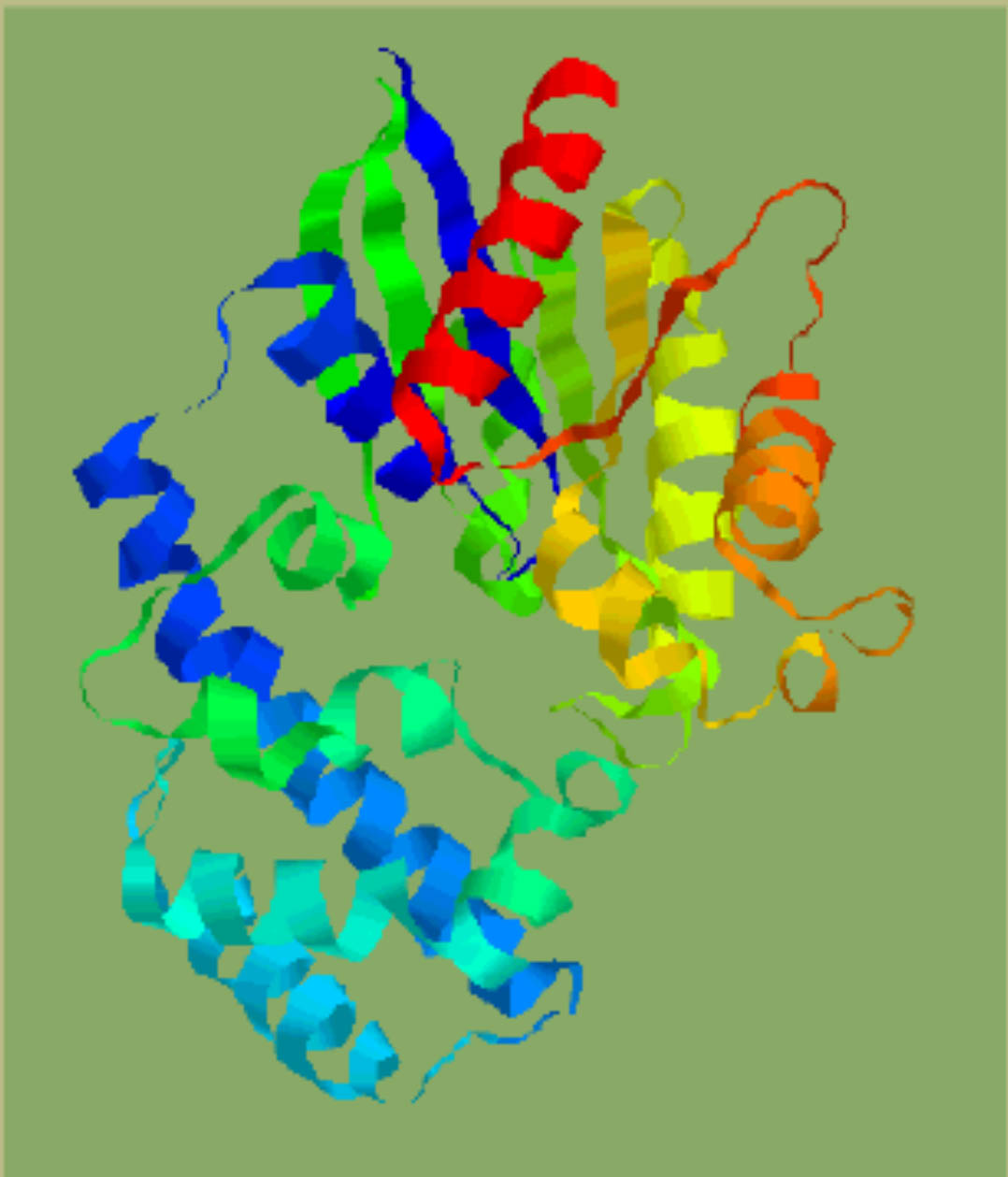
| Crystal structure of a-transducin. | Domain identification in a-transducin by clustering secondary structures. |
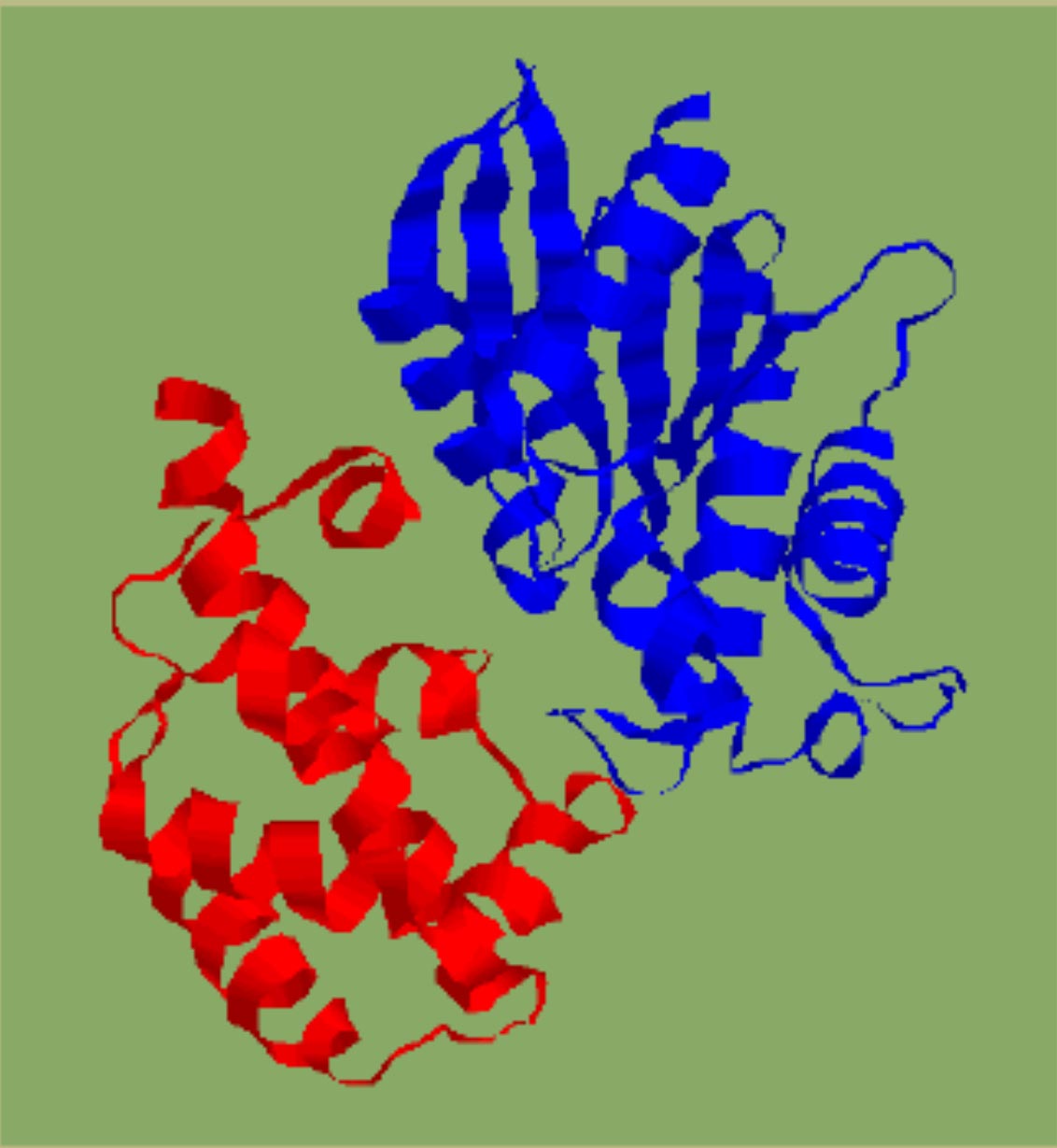 |
Identified domains in a -transducin ( N terminal discontinuous GTP-binding domain with the classical doubly-wound fold and a helix-rich insertion domain). |
2. Clustering of domains
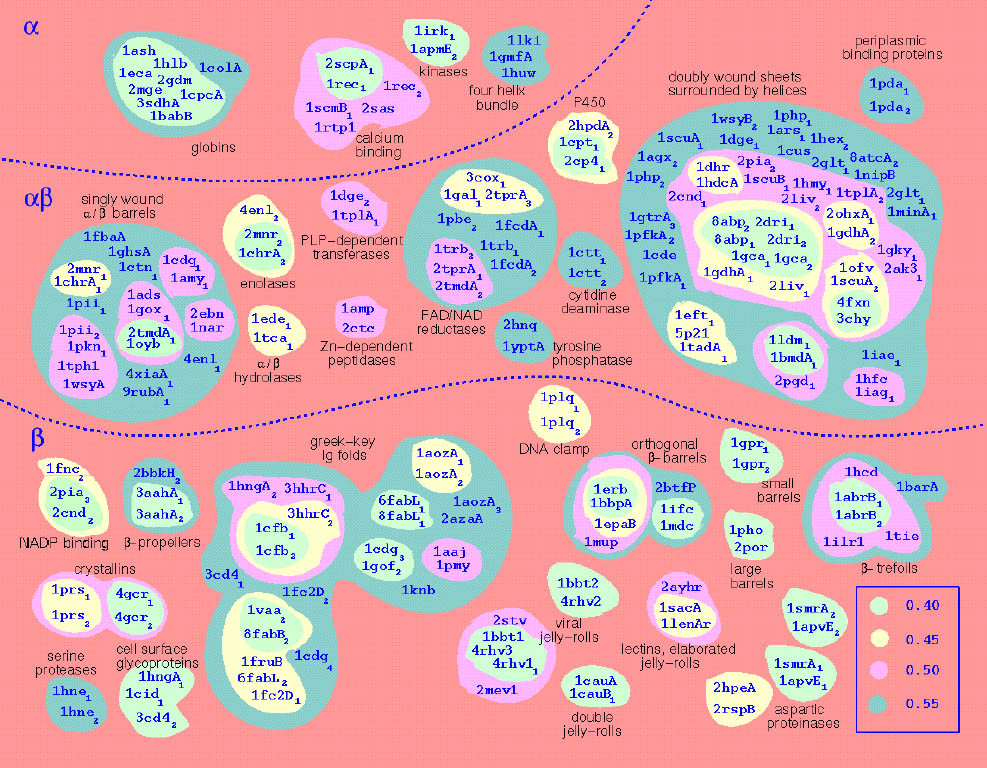 |
Structural similarities of 428 domains. |
|
|
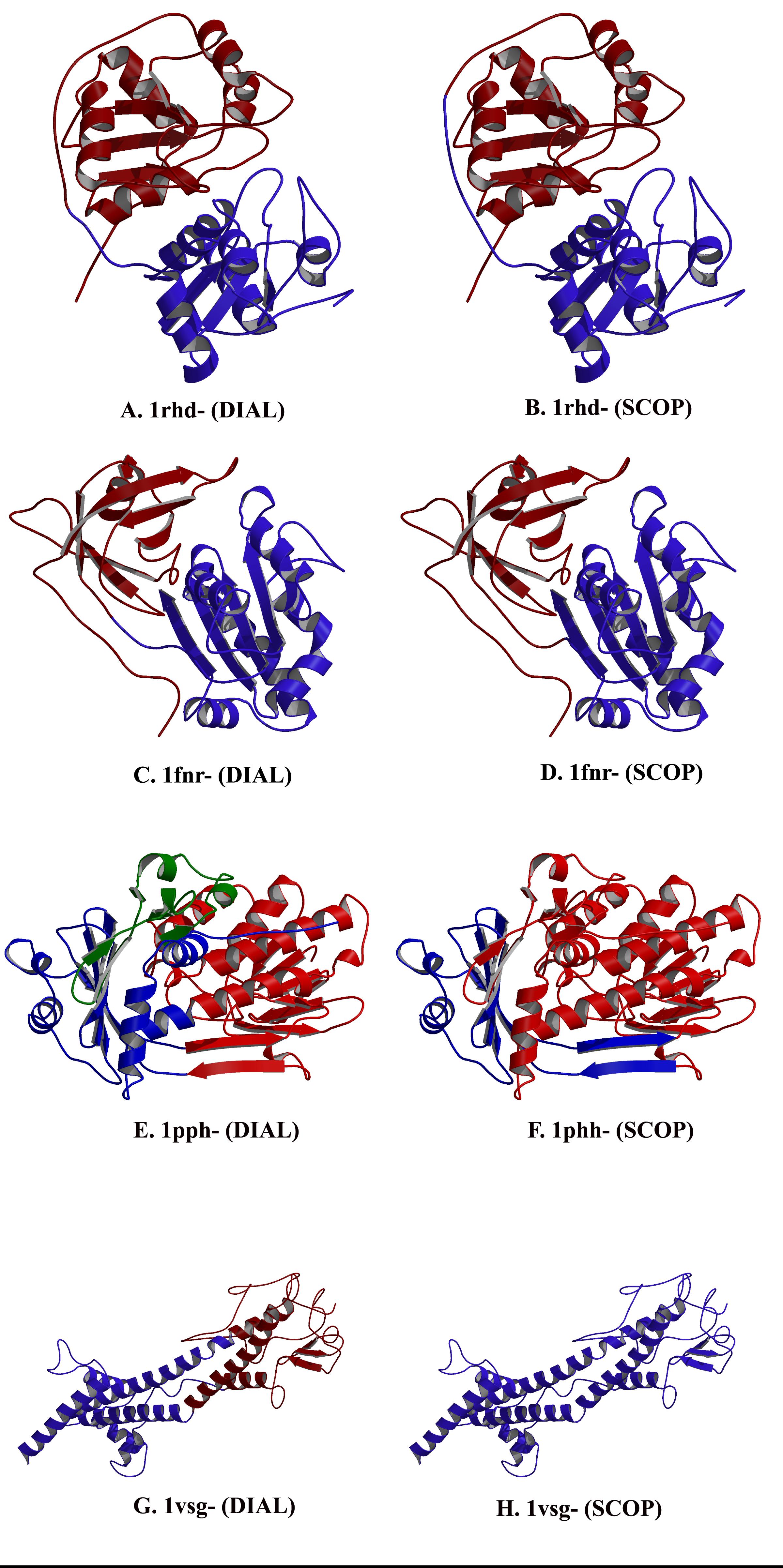 |
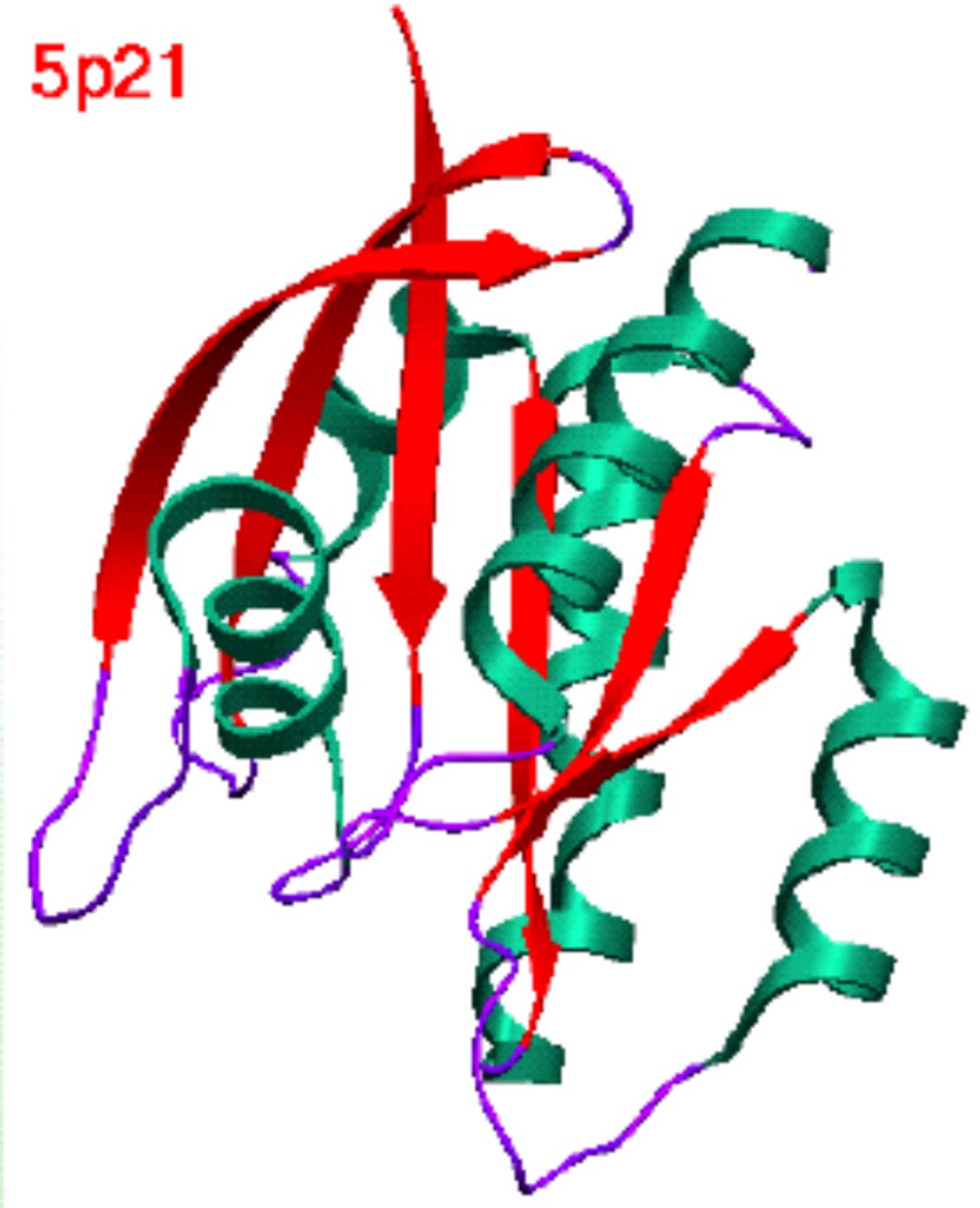
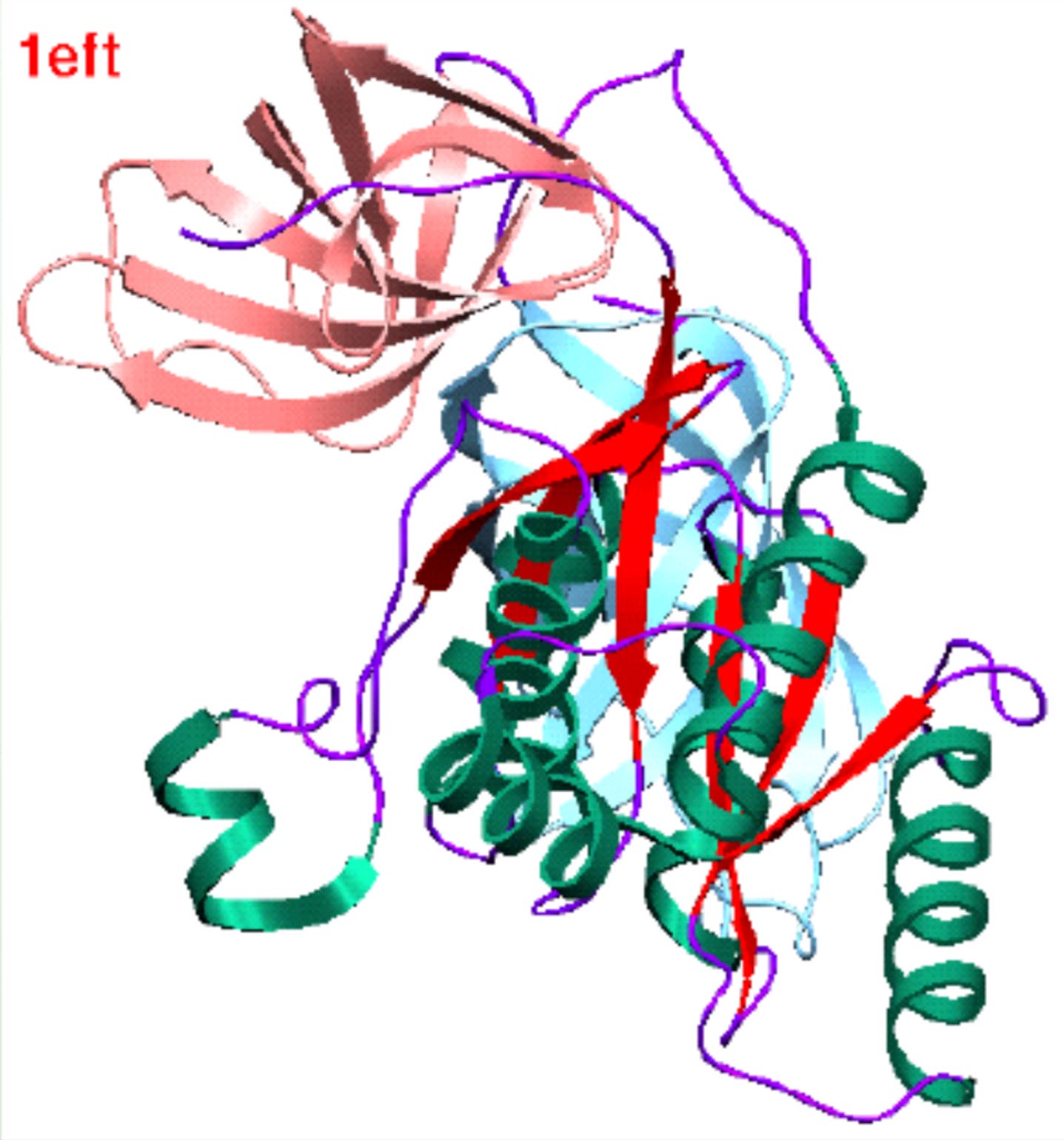
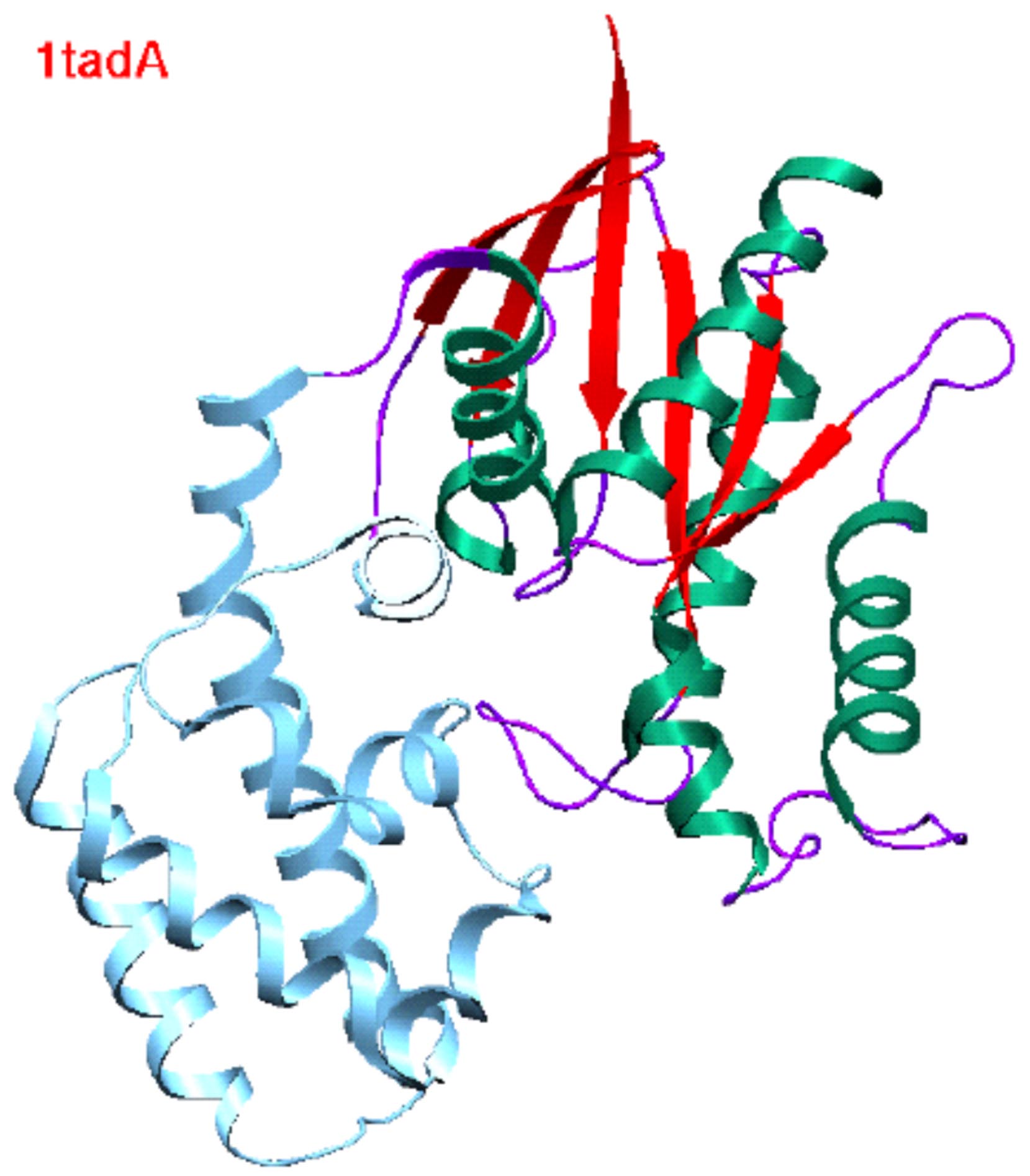
| Structure of proteins in the G-protein (ras) superfamily. | Domain definition by DIAL and SCOP. |
3. Model of the tetrameric arrangement of the transmembrane domain of inositol-triphosphate receptor
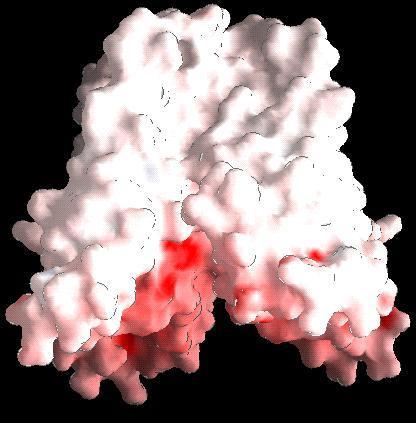 |
Model of the tetrameric arrangement of the transmembrane domain of inositol-triphosphate receptor. The model of the individual protomers have been obtained from the coordinates of the potassium channel (Doyle et al., 1998). Figure prepared by Parantu Shah |
4. Conserved and class-specific residues for TGF dimer and the TGF type-II receptor
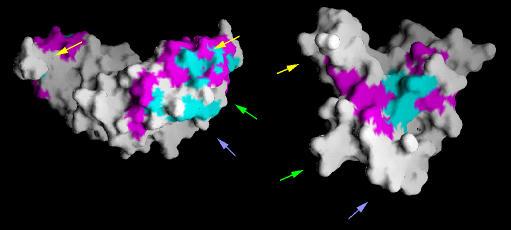 |
Conserved (shown in blue) and class-specific (shown in pink) residues shown for TGF dimer (left) and the TGF type-II receptor(right).Yellow, green and blue arrows indicate complimentary patches of residues that are predicted to be present at the interface regions. Figure prepared by Parantu Shah |
5. Models of members of bioluminescence superfamily - EagI model compared with LasI model
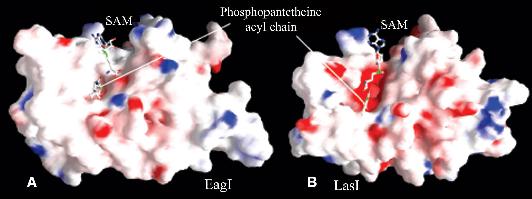 |
Models of members of bioluminescence superfamily - EagI model compared with LasI model. SAM and the corresponding PNT substrate have been docked and shown inball-and-stick form.Hydrophobic residues are shown in white, acidic residues in red and basic residues in blue.The PNT-binding site of LasI is broader and is acidic. Figure prepared by Saikat Chakrabarti |
6. Three-dimensional model of Bdm1 obtained by threading the sequence on the fold of BPHD
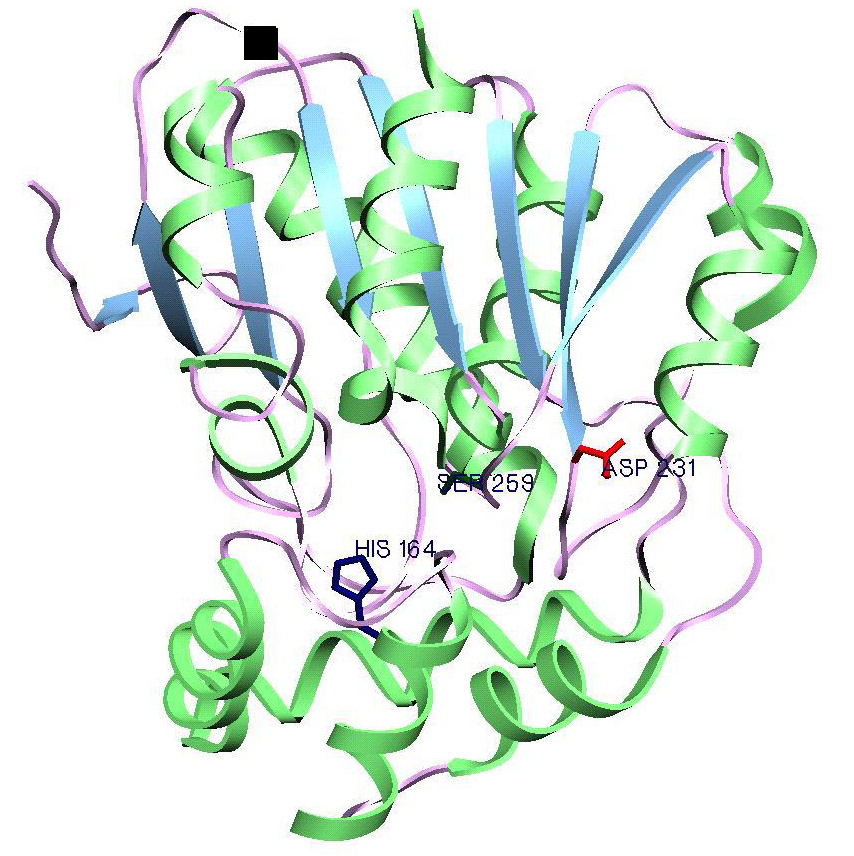 |
Three-dimensional model of Bdm1 obtained by threading the sequence on the fold of BPHD (pdbcode: 1c4x ) . Side-chains of three residues (His164, Ser259 and Asp231, identified by the functional template approach) shown qualify the criteria for the catalytic triad of alpha/beta hydrolases. Figure prepared by Anirban Bhaduri |
7.Disulfide bonds in AGITOXIN
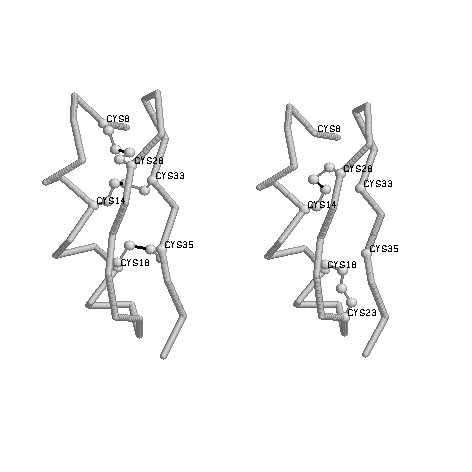 |
Fig 1:Native disulfide bonds of 1agt (annotated in PDBfile) Connectivities: 8-28,14-33,18-35 Figure prepared by Ratna Rajesh.T |
8.Structural Motifs in Globin-like superfamily
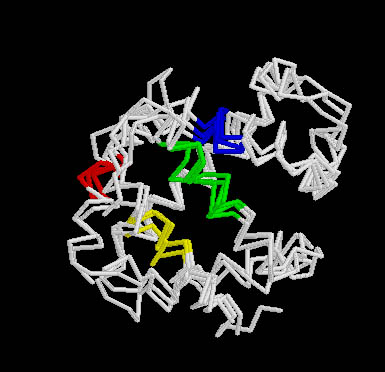 |
Orientation of structural motifs in Globin-like superfamily. Figure prepared by Saikat Chakrabarti |
9.Beta Barrel structure in alpha-hemolysin(7ahl) from staphylococcus aureus
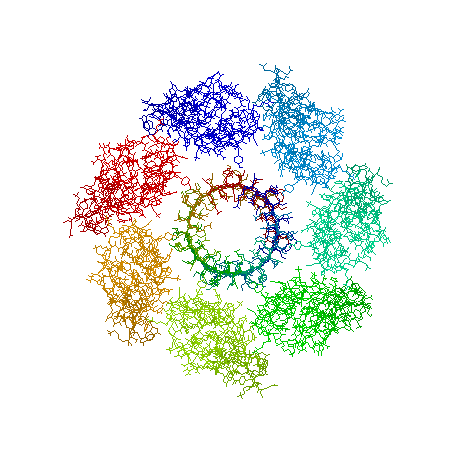 |
Beta Barrel structure in alpha-hemolysin (7ahl ) from staphylococcus aureus. Figure prepared by Pugalenthi.G |