Prof. R. Sowdhamini - Analysis of structure based sequence alignments
Homologous proteins resemble each other in sequence, structure and usually function. In general, proteins which share over about 30% sequence identity belong to a homologous family (Rossmann & Argos, 1977; Richardson, 1981; Chothia, 1984; Overington et al., 1993). Although the aligned sequences usually have small insertions and deletions, the local structural environment - solvent accessibility, secondary structure and sidechain hydrogen bonding - is conserved for residues at topologically equivalent positions (Rossmann & Argos, 1977; Overington et al., 1990; 1993). Divergent relationships undoubtedly also occur beyond the 'nuclear' family (Rossmann et al., 1974; Matthews & Rossmann, 1975) but they can be difficult to identify on the basis of sequences alone (Murzin & Chothia, 1992; Holm et al., 1992; Alexandrov & Go, 1994; Orengo et al., 1994; 1997; Sowdhamini et al., 1996; Holm & Sander, 1998; Russell et al., 1998; Swindells et al., 1998; Sternberg et al., 1999).
Distant relationships between proteins are often reflected in terms of their overall retention of structural similarity and high divergence at the sequence level. Few such distant relationships extend to similarities in function and therefore can be linked in evolutionary terms (also termed as superfamilies), whereas a majority of proteins which look similar perform different functions. Proteins which can be grouped into superfamilies share a common fold, may have similar packing of secondary structural elements, often have similar functions, but have very little sequence similarity (Overington et al., 1990; Rossmann & Argos, 1977; Holm et al., 1992; Alexandrov & Go, 1994; Orengo et al., 1994; Sowdhamini et al., 1996; Murzin et al., 1995; Martin et al., 1998; Castillo et al., 1999; Orengo et al., 1999). Such distant relationships between proteins pose challenges in the prediction of fold and function of new proteins for which only sequence information is available. Structural similarity often exists at the level of protein domains (Wetlaufer, 1973; Richardson, 1981; Wodak & Janin, 1981; Go, 1981).
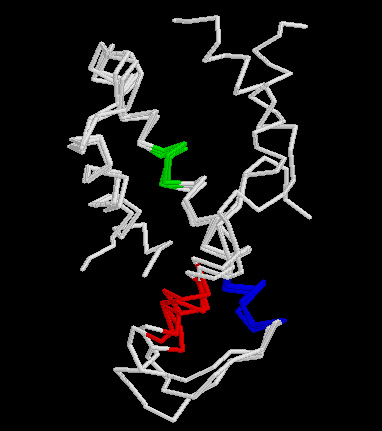
Superfamily is a hierarchical classification that contains proteins of different families and subfamilies having similar structure and function. These proteins might have very low sequence identities but retain the same fold through well-conserved secondary structural parts. On the basis of conservation of criteria, like amino acid preference and solvent accessibility, several conserved segments of proteins belonging to the same superfamily have been identified. These segments are termed as 'structural motifs'. These motifs, along with their sequence and spatial orientation and preservation of various structural criteria, represent the conserved core of each superfamily. The structural features of such motifs for several superfamilies have been integrated into the SMoS database. The definition of superfamilies is in direct correspondence with SCOP (Murzin et al., 1995). One of the main purposes of SMoS is to provide important sequence segments that can be projected as the minimum structural requirements for a new member to be considered part of a pre-existing superfamily. Such motifs can also be employed to design and rationalize protein engineering and folding experiments.
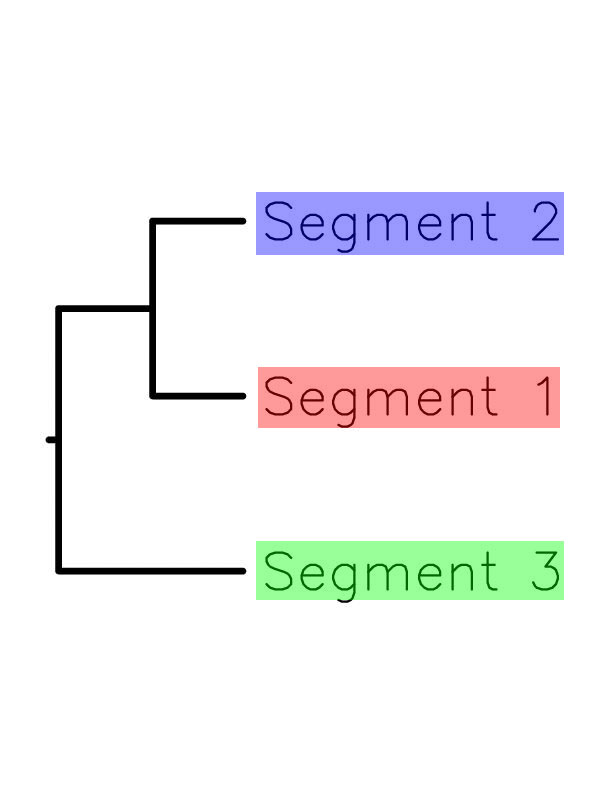
| Structural motifs of Histone-fold superfamily | Tree file |
 |
 |



