Dr. Vinothkumar Kutti Ragunath - Research Area
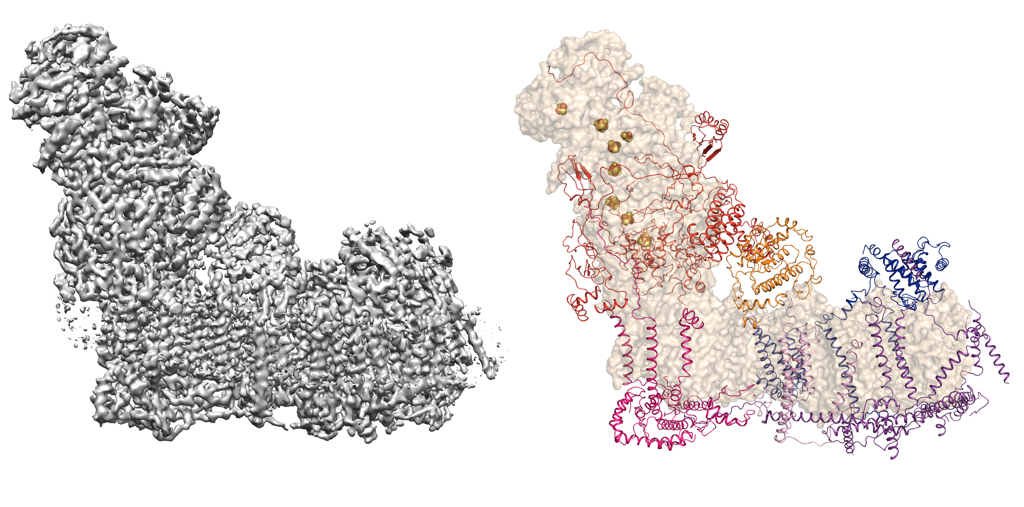
The hydrophobic nature of membrane proteins presents several unique challenges in their structure determination including their low abundance or difficulty in over-expression of functional material and stability in detergents. Some of these issues can be mitigated with use of single particle electron cryomicroscopy (cryoEM) as there is no need to grow crystals and very little protein is required. Recent advances in cryoEM has resulted in structure determination of number of macromolecules, which otherwise were very difficult to study structurally. These molecules can be now be imaged and high-resolution structures can be obtained with very little protein required. The wonderful thing about EM is that one can visualise the protein of interest on the micrographs to obtain immediate feedback on the quality, thus the biochemical preparation can be improved. One such example where EM helped in structure determination is the bovine Complex I from mitochondrial inner membrane. Complex I in higher eukaryotes is composed of 45 subunits, many of which are encoded in nucleus, imported into the mitochondria and assembled. Many of these subunits are membrane proteins and the complex has a total of 78 transmembrane helices. The figure below shows an EM map of Complex I, reconstructed from the enzyme solubilised in detergent micelles and the model that was built into the map thus allowing us to assign all subunits (Vinothkumar et al 2014; Zhu et al 2016).
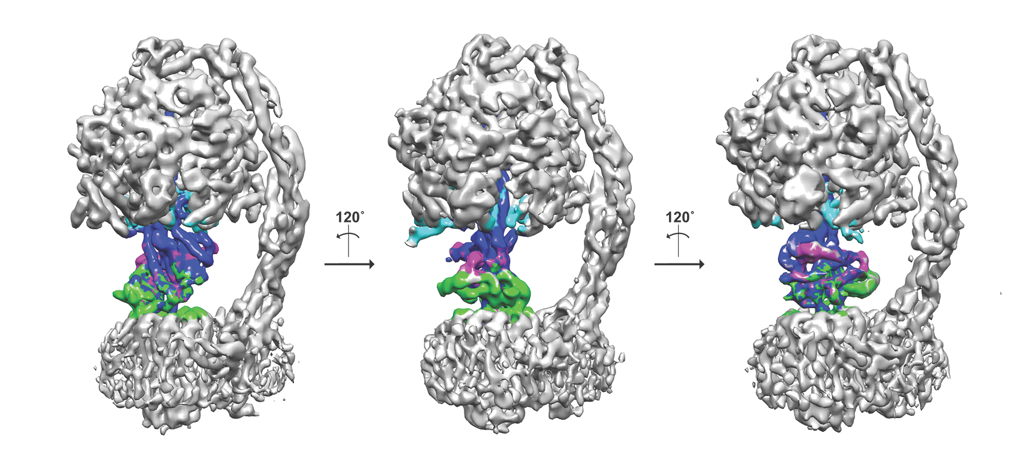
Another added advantage is that the freezing process can trap macromolecules in different functional states, multiple structures and thus the dynamics of a given protein can be studied. The mitochondrial F1Fo-ATP synthase from Pichia angusta in three different states is shown as an example here. ATP synthase's are dynamic enzymes that use the proton/sodium gradient to synthesise ATP and can exist in multiple different states. Using a protein inhibitor found in mitochondria, the enzyme has been trapped in different states and computationally these states can be separated from a mixture of population. The central stalk that consists of three proteins is coloured in different colours (blue, magenta and green) and the inhibitor protein in cyan. The three states are related roughly by 120˚ rotation (Vinothkumar et al., 2016)
My research interest is thus focused on understanding the struture and mechanism of membrane proteins using cryoEM as the main technique but also X-ray crystallography and other biochemical and biophysical techniques. The family of membrane proteins that we are currently pursuing include membrane enzymes, ion channels and multisubunit protein complexes.



