Dr. Mrinalini Puranik - Research Report 2004-2005
Research Report 2004-2005 :
Structure and dynamics of biomolecules
Our broad interest is to understand the structure, reactivity and dynamics of proteins and nucleic acids, especially in the context of DNA-molecule and DNA-protein interactions involved in damage and repair processes. In the cellular environment, DNA can react with endogenous and exogenous molecules leading to damage via covalent modification of the bases and ribose and protein-DNA cross linking. While the end products of these damaging reactions are known, the chemical mechanisms are not understood. In the presence of these reactions, maintaining genomic integrity is critical to all living organisms. Many complex but efficient pathways have evolved in eukaryotes and prokaryotes to rectify these modifications. The pathway involved in repair of modified single bases is called base excision repair (BER). Enzymes involved in BER and their substrates have been identified but the precise nature of the interaction of the base with the enzyme and subsequent catalytic repair is not understood. Questions of intense current interest pertain to the mechanisms of recognition of damage sites, high substrate specificity and chemical process of repair.
We aim to obtain a molecular level understanding of both the damage and repair processes. To elucidate the mechanism of DNA damage we are identifying and characterizing the complexes and reactive species of bases and damage agents formed on photoexcitation. This is challenging since reactive species have typical lifetimes on the order of nanoseconds. The subsequent enzymatic repair process by a specific set of enzymes, the glycosylases involved in BER pathway will also be extensively studied.
To investigate these issues, we have setup a facility for ultra violet resonance Raman spectroscopy (RRS). RRS yields information on the structure of molecules through their vibrational spectra. A unique feature of this technique is that normal and modified nucleobases can be selectively observed without intereference from the rest of the solution and interacting proteins. Using a combination of RRS with site-directed mutagenesis, we expect to gain insight into the detailed mechanisms of damage, recognition and repair.

|
Ultra-violet resonance Raman facility at NCBS
|
1. Mechanisms of photo-induced DNA damage
The DNA damage potential of high energy ultraviolet rays (UVB and UVC) is known for a long time. That the lower energy UVA radiation is also damaging is demonstrated by recent work. Endogenous and exogenous chromophores like porphyrins and flavins are potential genotoxic agents when photoexcited e.g., riboflavin (RF). While the end products of the reactions of damage agents and DNA are known, the chemical mechanisms are not well understood. Identifying the reaction intermediates will provide a detailed insight into these reactions.
Photoexcited RF preferentially oxidizes the 5’-G of a –GG– repeat to form 8-oxo-dG. The intermediates of RF and dGMP involved in the reaction are not known since biochemical assays can only detect the end product of the reaction. The 8-oxo-dG lesion is especially important since it leads to transversion mutations of G:C to T:A and is more reactive than guanine. The aim is to observe the formation of these lesions with nanosecond time resolution after initiation of the photoreaction. In a two laser experiment, one laser will be used to photoexcite the RF molecule and the second laser will be used to observe the formation of guanine and RF intermediates as the reaction proceeds. Steady state RRS experiments and quantum chemical calculations are in progress to obtain the spectral signature of 8-oxo-dG and characterize the possible RF-DNA complex and photoproducts.
2. Enzymatic DNA repair by glycosylases
The repair process involves specific DNA-protein interactions in which the damaged
lesions are identified, excised and the original sequence is restored. It is estimated that the BER pathway is involved in the repair of 20,000 bases per cell per day. Formadopyrimidine glycosylases (Fpg), the first enzymes to be recruited in the BER pathway excise singlebase DNA lesions and create an apurinic site. The damaged base 8-oxo-dG differs from the normal base, guanine by only two atoms. Thus, the mechanism of recognition and substrate specificity of FPG has generated a keen interest.
Recent crystal structures of Fpg and Fpg-DNA complexes reveal the residues in the
binding pocket. Details of the mechanisms of recognition and catalysis can be determined unequivocally by directly probing the damaged nucleobase bound to Fpg in solution. WT and site-directed mutants of r esidues in the active site pocket of Fpg will be made and observed by probing in resonance with the nucleobase and protein in independent experiments.
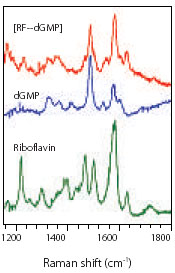 |
Figure 1: Typical ultra-violet resonance Raman spectra of dGMP and Riboflavin and their ground state complex in phosphate buffer at pH 7.0. Spectra were obtained with UV excitation at 267 nm and 0.4 mW incident power. |
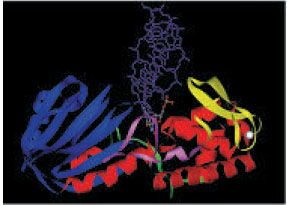
| Figure 2: Picture shows a variant of E. coli Fpg (E3G) bound to dsDNA containing an 8-oxo-dG lesion flipped out into the active site (PDB file: 1R2Y). The 8-oxo-dG lesion is shown in ball-and-stick model and only one of the DNA strands is shown in purple. The DNA binding site is a groove between the N- (blue) and C-termini (red). Several residues forming a loop of the protein (pink) make contacts with the lesion. The zinc-finger region is shown in yellow and the zinc atom in white. |
 |



