Dr. Yamuna Krishnan
Moved to University of Chicago
Our group is moving to the Department of Chemistry, University of Chicago.
We have had a GREAT time at NCBS, a very special place indeed and continue to cherish our strong ties here.
Structure and Dynamics of Nucleic Acids
Bionanotechnology aims to learn from nature - to understand the structure and function of biological devices and to utilise nature's solutions in advancing science and engineering. Evolution has produced an overwhelming number and variety of biological devices that function at the nanoscale or molecular level. My lab’s central theme is one of ‘synthetic biology’, which involves taking a biological device, component or concept out of its cellular context and harnessing its function in a completely new setting to probe, program or even reprogram living systems. Our current research involves understanding the structure and dynamics of unusual forms of DNA and translating this knowledge to create DNA-based nanodevices for applications in bionanotechnology.
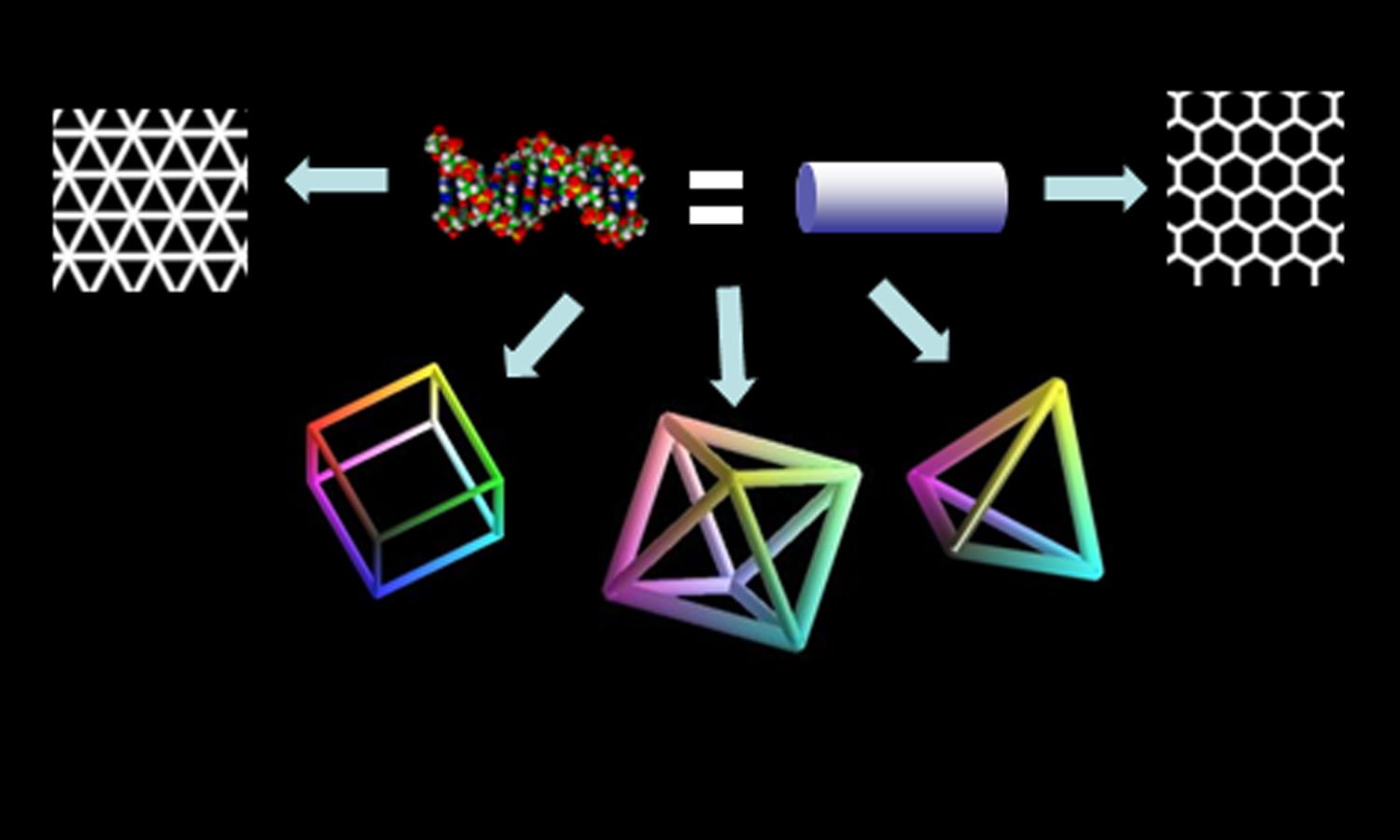
Structural DNA nanotechnology is an emerging field that uses the base-complementarity design principle of DNA to create ordered superstructures from a set of DNA sequences that self-assemble into regular, well-defined topologies on the nanoscale. With a diameter of 2 nm and a helical periodicity of 3.5 nm, the DNA double helix is inherently a nanoscale object. The specificity of Watson-Crick base pairing endows oligonucleotides with unique and predictable recognition capabilities. This makes DNA an ideal nanoscale construction material. Understanding and thereby controlling structure and dynamics in DNA is thus key to realizing its potential as a nanoscale building block for device applications of structural DNA nanotechnology. These DNA nanodevices may function as rigid scaffolds in 1D, 2D or 3D or as dynamic switches.
Selected Publications:
- Sharma, S., Zaveri, A., Visweswariah, S.S and Krishnan, Y.* (2014) A fluorescent nucleic acid-based nanodevice quantitatively images elevated cAMP in membrane bound compartments. Small, in press.
- Chakraborty, S., Mehtab, S. and Krishnan, Y.* (2014) The predictive power of synthetic nucleic acid technologies in RNA biology. Acc. Chem. Res.47, 1710-1719.
- Modi, S., Nizak, C., Surana, S., Halder, S. and Krishnan, Y.* (2013) Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nature Nanotechnology, 8, 459-467.
- Banerjee, A., Bhatia, D., Saminathan, A., Chakraborty, S., Kar, S. and Krishnan, Y.* (2013) Controlled release of encapsulated cargo from a DNA icosahedron using a chemical trigger. Angew. Chem. Int. Ed. 52, 6854-6857.
- Chakraborty, S., Mehtab, S., Patwardhan, A.R., Krishnan, Y.* (2012) Pri-miR-17-92a Transcript folds into a tertiary structure and autoregulates its processing. RNA 18, 1014-1028.
- Bhatia, D., Surana, S., Chakraborty, S., Koushika, S.P. and Krishnan, Y.* (2011)A synthetic, icosahedral DNA-based host-cargo complex for functional in vivoimaging. Nature Communications, 2, 339
If you've reached this far, see some features on our work! http://www.nature.com/nindia/2013/130528/full/nindia.2013.69.html http://www.nature.com/nnano/reshigh/2011/0611/full/nnano.2011.91.html http://www.nature.com/nindia/2011/110629/full/nindia.2011.99.html http://www.financialexpress.com/news/giving-dna-nanodevices-a-new-role-inside-living-systems/875447/0