Demo Simulation :
On these pages we present complete simulations to supplement selected publications. These simulations are usually in the form of 'demonstration' simulations that will run with minimal user involvement. The demonstrations typically generate figures reported in our publications and are intended to make it absolutely explicit how the simulation results were obtained. There are also numerous supplementary simulations that for reasons of space were not incorporated into the print publications, but may be valuable for further understanding the model and its biological implications.
The demonstration simulations run with GENESIS and Kinetikit.
Although Kinetikit is designed as a friendly graphical interface, its data storage format is somewhat difficult. Users who wish to use the parameters in other contexts are referred to the DOQCS database, where these and many other signaling models are stored in a more accessible manner.
Selected publications with demonstration simulations and supplementary material:
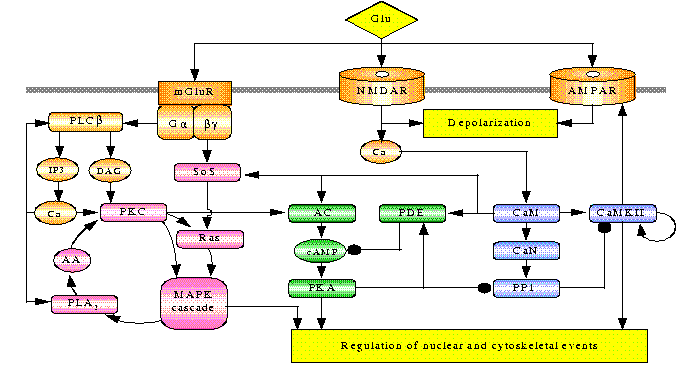 |
U.S. Bhalla and R. Iyengar,
Science 283:381-387 (1999)
Emergent Properties of Networks of Biological Signaling Pathways.
|
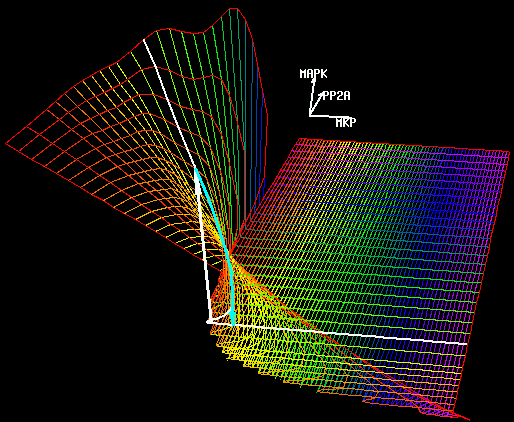 |
U.S. Bhalla, P.T. Ram and R. Iyengar,
Science 297:1018-1023 (2002)
MAP Kinase Phosphatase As a Locus of Flexibility in a Mitogen-Activated Protein Kinase Signaling Network.
|
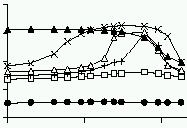 |
U.S. Bhalla,
Biophys.J. 83(2):740-752, 2002.
Mechanisms for temporal tuning and filtering by postsynaptic signaling pathways.
|
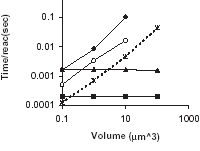 |
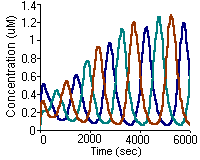
K. Vasudeva and U.S. Bhalla
Bioinformatics 20(1):78-84, 2004.
Adaptive stochastic- deterministic chemical kinetic simulations.
|
 |
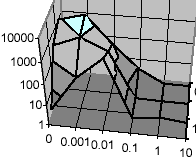
S.J. Vayttaden and U.S. Bhalla
Sci STKE. 2004 Feb 3;2004(219):pl4.
Developing complex signaling models using GENESIS/Kinetikit.
|
 |
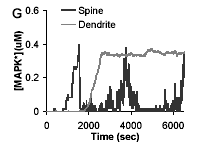
U.S. Bhalla
Biophys. J. 87(2):733-744, 2004.
Signaling in small subcellular volumes. I. Stochastic and diffusion effects on individual pathways.
|
 |
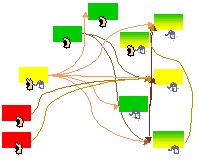
U.S. Bhalla
Biophys. J. 87(2):745-753, 2004.
Signaling in small subcellular volumes. II. Stochastic and diffusion effects on synaptic network properties.
|
 |
S.J. Vayttaden, S.M. Ajay and U.S. Bhalla
ChemBioChem 2004 5(10):1365-1374
A Spectrum of Models of Signaling Pathways.
|
 |
D.K. Sinha, U.S. Bhalla, Shivashankar, G.V.
App. Phys. Lett 2004. 85(20):4789-4791
Kinetic measurement of ribosome translation force.
|
 |
G.V. HarshaRani, S.J. Vayttaden, U.S. Bhalla
Journal of Biochemistry 2005. 137(6)
Electronic data sources for kinetic models of cell signaling.
|
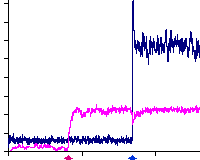 |
A. Hayer, U.S. Bhalla
PLoS Computational Biology 2005. 1(2):e20
Molecular switches at the synapse emerge from receptor and kinase traffic.
|
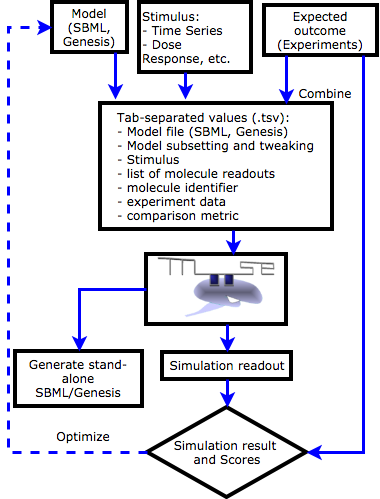 |
FindSim:a Framework for Integrating Neuronal Data and Signaling Models
|















