Prof. Upinder S. Bhalla - Ribosome translational forces Demo
Supplementary information and demonstration simulations for:
Kinetic measurement of ribosome translation force
(App. Phys. Lett 2004. 85(20):4789-4791)
Authors:
Sinha, D.K.: National Center for Biological Sciences
Bhalla, U.S. National Center for Biological Sciences and
Shivashankar, G.V.* National Center for Biological Sciences
*Corresponding author.
Models of gene expression.
Downloading GENESIS and Kinetikit.
Database of signaling pathway models.
Supplementary material 1. (pdf file)
Supplementary material 2. (doc file)
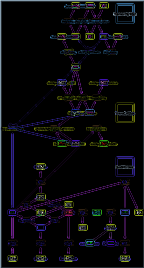
Click to see full-screen dump of ribosome translation model
Abstract
We measure the ribosomal translation forces to unzip mRNA polymers during gene expression. A novel approach of using the changes in the reaction rate constants to determine the molecular forces is presented. Specific antisense DNA oligomers complementary to mRNA templates are used as kinetic barriers for estimating the ribosome forces using real time biolouminescence detection of luciferase gene expression. We measure for the first time the translation kinetics using fluorescence anisotropy, to show that the antisense DNA oligomers only affect the translation kinetics. The rate constants are determined by comparing the experimental data with numerical simulation of gene expression to deduce the ribosome force (26.5 +-1 pN) required to unzip mRNA polymers. Our data may have direct implications in understanding the physical mechanisms that underlie translation regulation.



